Documentation
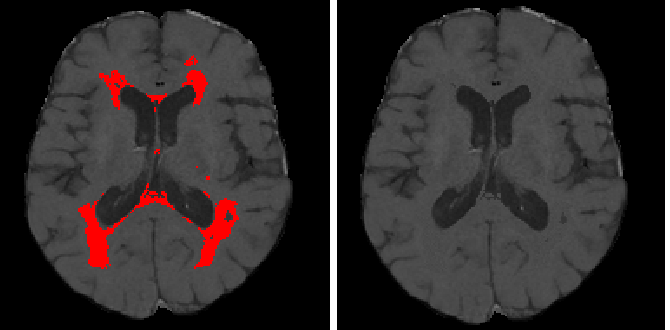
Lesion Filling
Our lesion filling has been implemented using the Matlab environment. The method has been implemented both as a Matlab function, and as a SPM external library. Here we detail the necessary steps to install SLF.
Stand-alone function
The installation is straightforward. Download the toolbox and extract it into a folder. Then, just add the selected folder into the Matlab path.The function SLF(input_scan, output_scan, lesion_mask) saves a NIFTI images on the output_scan location provided as input. All input parameters should be existing paths to NIFTI images. The function assumes that the input T1-w image is currently skull-stripped. For instance:
input_scan = '/dir/to/input/image/foo.nii'
refilled_scan= '/dir/to/output/image/output.nii'
lesion_mask= '/dir/to/lesionmask/image/foomask.nii'
SLF_lesion_filling(input_scan, refilled_scan, lesion_mask)
would save the refilled_image image on '/dir/to/output/image/output.nii'.
The function SLF(input_scan, output_scan, lesion_mask, brainmask) saves a NIFTI image on the output_scan location provided as input. All input parameters should be existing paths to NIFTI images. Contrary to the previous function, the brainmask is also provided. For instance:
input_scan = '/dir/to/input/image/foo.nii'
refilled_scan= '/dir/to/output/image/output.nii'
lesion_mask= '/dir/to/lesionmask/image/foomask.nii'
brain_mask = '/dir/to/brainmask/image/brainmask.nii'
SLF_lesion_filling(input_scan, refilled_scan, ...
... lesion_mask, brain_mask)
would save the refilled_image image on '/dir/to/output/image/output.nii'.
SPM library
Installing SLFtoolbox as a SPM library is also straightforward. Download the toolbox, and extract it into a folder. These are the required steps to run SLF inside the SPM bundle.
- On SPM, all external libraries are installed into the toolbox folder. This folder is located inside the main SPM directory. Just find where SPM8 was installed and open the toolbox folder.
- Download the toolbox, and extract it into the spm/toolbox folder.
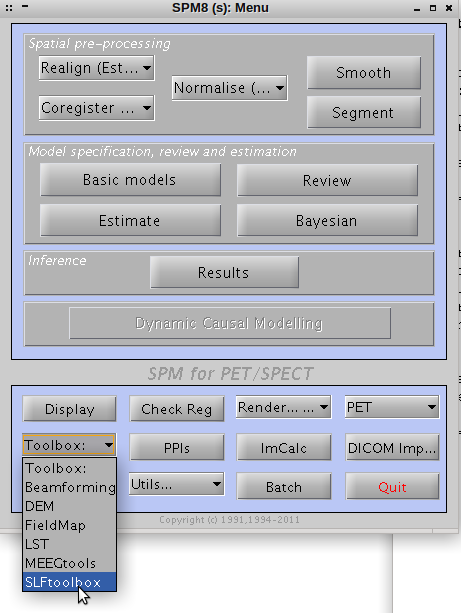
- Open MATLAB as usual, and type spm in the command-prompt.
- This will open the main SPM8 menu. Hence, on the Toolbox list, select the SLFtoolbox.
- The SPM batch editor will be opened containing the SLFtoolbox program.
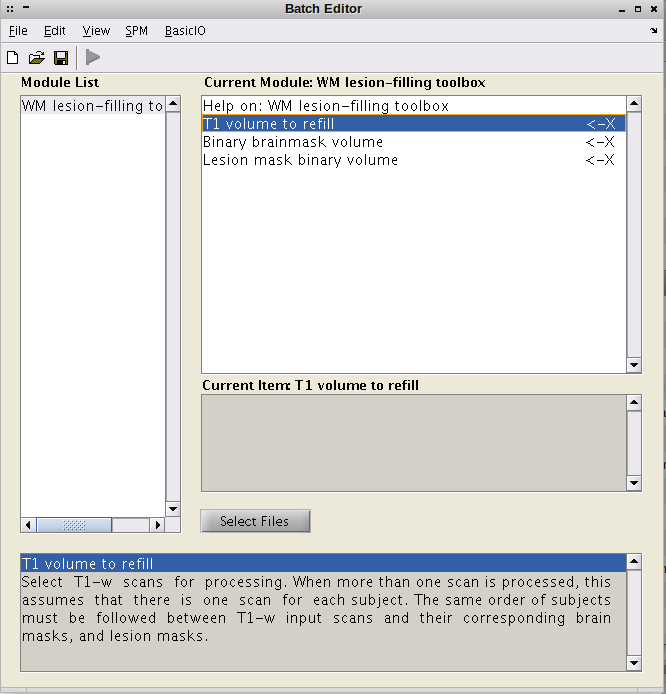
- A T1-w image to refill. If more of one image is selected, the same order must be followed for all T1-w images and their corresponding brain and lesion masks.
- A binary brainmask image. If more of one image is selected, these images must follow the same order entered for the T1-w images to refill.
- A binary lesion mask image. If more of one image is selected, these images must follow the same order entered for the T1-w images to refill.
Other lesion filling codes
The codes in this section are provided by its respective authors and all we do here is provide some pointers to make it easier to access it.
LEAP: Chard DT, Jackson JS, Miller DH, Wheeler-Kingshott, CA (2010): Reducing the impact of white matter lesions on automated measures of brain gray and white matter volumes. Journal of Magnetic Resonance Imaging 32: 223–228.
FSL Lesion FillingFSL-L: M. Battaglini, M. Jenkinson, and N. De Stefano. Evaluating and reducing the impact of white matter lesions on brain volume measurements. Human Brain Mapping, 33(9):2062–2071, 2012.